Abstract
Background
Chronic myelomonocytic leukemia (CMML) is a heterogeneous clonal disorder characterized by peripheral blood monocytosis and a propensity for acute myeloid leukemia (AML) transformation. CMML patients (pts) have a variable disease course that ranges from indolent to rapid progression and death.
Several prognostic scoring systems have been developed to risk stratify CMML pts including: IPSS, R-IPSS, Global MD Anderson Scoring System (MDASC), MD Anderson Prognostic Score (MDAPS), Dusseldorf Score (DS), Mayo prognostic model, and the CPSS. While a large study from the MDS/MPN International Working Group found that these models are valid and have comparable performances, even in pts with high white blood cell counts, considerable heterogeneity in their prediction of outcomes is observed when applied in clinical practice. Understanding this heterogeneity is clinically essential as treatment strategies can differ based on the model used and the expected outcomes.
Method
Clinical data from pts who were diagnosed with CMML according to 2008 WHO criteria between 7/1981 and 7/2014 was abstracted from 4 institutions in the US. All prognostic models were calculated as previously described. Overall survival (OS) was calculated from the time of diagnosis to time of last follow up or death. Descriptive statistics were used to study the changes in risk categories across models.
Result
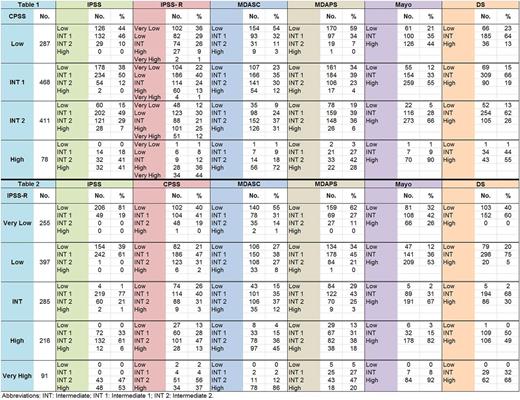
Of 1244 pts included in the analysis, the median age at diagnosis was 70 (16-91) years with a male 67% predominance. By WHO classification, the majority of pts had CMML-1 (79.9% vs. 20.1%) and 50% had MPN-CMML by FAB criteria. Among 755 pts with low-risk (low / INT-1) disease per CPSS, 11% were reclassified as higher-risk (defined as INT-2 or high risk) by IPSS, 12% by IPSS-R, 31% by MDASC, 19% by MDAPS, and 34% as INT and 51% as high by Mayo, Table 1. Among 489 pts with higher-risk disease (INT-2/ High) by CPSS, 56% were reclassified as lower-risk by IPSS, 56% by IPSS-R, 29% by MDASC, 53% by MDAPS, and 5% as low and 25% as INT by Mayo, Table 1.
Among 652 pts (52%) with very low/low risk disease per IPSS-R, 20% were re-classified as INT-2 and 5% as high-risk by MDASC, 17% as INT-2 by MDAPS, 38% as INT and 42% as high by Mayo, and 26% as INT-2 and 1% as high by CPSS, Table 2. Among 285 pts with INT risk per IPSS-R, 78% were reclassified as lower-risk by IPSS, 50% by MDASC, 72% by MDAPS, 66 % by CPSS, and 67% were re-classified as high risk by Mayo, Table 2. Among 307 pts with high/very high-risk per IPSS-R, 2% were reclassified as lower-risk by IPSS, 21% by the MDASC, 35% by MDAPS, 30% by CPSS, and 13% as INT by Mayo, Table 2.
When calculating the difference between the predicted OS and the actual OS (evaluable in 834 pts) by a given model, almost all models overestimated the OS of pts in lower-risk categories. The difference between the median predicted OS and median actual OS was 43.7 months (m) for low risk, 22.5 m for INT-1, 0.38 m for INT-2 and -4.1 m for high risk (pts lived longer than their predicted OS) by IPSS , 58.7 m for very low/low, 18.78 m for INT, 5.0 m for high and 2.27 m for very high by IPSS-R, 72.4 m for low, 38.5 m for INT-1, 21.2 m for INT-2 and 11.0 m for high by CPSS, and 6.47 m for low, -6.93 m for INT, and -3.95 m for high risk by Mayo.
Conclusion
Significant intra-patient (difference of outcome in the same pt based on the model used) and intra-group (difference in outcome among pts who have similar risk category by a given model) variation is observed when applying commonly used models in CMML. Further, significant differences exist between the predicted OS and the actual OS of pts across all models. Almost all models overestimated the OS of pts within lower-risk categories. Adding molecular data especially ASXL1 mutations may help risk stratify these pts more effectively.
Over-estimation of survival using these models may set unreasonable expectations regarding the disease course and treatment recommendations. A personalized prediction model that can precisely predict the actual outcome of a pt is warranted.
Komrokji: Novartis: Honoraria, Speakers Bureau; Celgene: Honoraria. Daver: Immunogen: Research Funding; Karyopharm: Consultancy, Research Funding; Novartis Pharmaceuticals Corporation: Consultancy; Bristol-Myers Squibb Company: Consultancy, Research Funding; Jazz: Consultancy; Kiromic: Research Funding; Pfizer Inc.: Consultancy, Research Funding; Incyte Corporation: Honoraria, Research Funding; Otsuka America Pharmaceutical, Inc.: Consultancy; Daiichi-Sankyo: Research Funding; Sunesis Pharmaceuticals, Inc.: Consultancy, Research Funding. Savona: Gilead: Membership on an entity's Board of Directors or advisory committees; Sunesis: Research Funding; Incyte Corporation: Consultancy, Research Funding; Astex: Membership on an entity's Board of Directors or advisory committees, Research Funding; Karyopharm: Consultancy, Equity Ownership; Amgen: Membership on an entity's Board of Directors or advisory committees; Celgene: Membership on an entity's Board of Directors or advisory committees; Takeda: Research Funding; TG Therapeutics: Membership on an entity's Board of Directors or advisory committees, Research Funding. Itzykson: Novartis: Research Funding; Janssen: Research Funding. Fenaux: Celgene: Honoraria, Research Funding; Amgen: Honoraria, Research Funding; Astex: Honoraria, Research Funding; Novartis: Honoraria, Research Funding; Celgene: Honoraria, Research Funding; Novartis: Honoraria, Research Funding; Janssen: Honoraria, Research Funding; Janssen: Honoraria, Research Funding; Astex: Honoraria, Research Funding; Amgen: Honoraria, Research Funding. Santini: Novartis: Honoraria; Abbvie: Consultancy, Membership on an entity's Board of Directors or advisory committees; Celgene: Honoraria, Research Funding; Janssen: Consultancy, Honoraria; Otsuka: Consultancy; Amgen: Membership on an entity's Board of Directors or advisory committees. Sekeres: Celgene: Membership on an entity's Board of Directors or advisory committees. Padron: Incyte: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal